30 Fitting Machine Learning Models
-
The package
scikit-learnincludes a comprehensive tools to conduct predictive analysis in machine learning framework -
The package is built on other packages like
NumPy,SciPyandMatplotlib -
It includes modelling option for both supervised and unsupervised problems
-
Methodologies include classification, regression, clustering, dimension reduction
-
It also includes several supportive tools likes model selection, preprocessing, feature extraction etc.
30.1 Steps of model fitting
Import specific modules of
scikit-learnImport other relevant libraries:
pandas,numpyRead the
pandasDataFrameSummary information of the dataset
Create training and testing datasets
Exploratory analysis of the training dataset
Build and fit the model
Evaluate the model on the test dataset
Explore the model performance
Use the model to predict unseen dataset
30.2 Load Libraries & Read Data 
Import scikit-learn
Note: We are importing modules for building a Decision Tree classifier
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn import metrics
from sklearn.tree import DecisionTreeClassifier, plot_tree
Import other libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
Set the working directory to the data folder
Read the iris dataset as a Python object DF
DF = pd.read_csv('iris.csv')
30.3 Data Summary 
Note: The code demonstrates only a selective summary
# Describe the data
DF.describe() SepalLength SepalWidth PetalLength PetalWidth
count 150.000000 150.000000 150.000000 150.000000
mean 5.843333 3.057333 3.758000 1.199333
std 0.828066 0.435866 1.765298 0.762238
min 4.300000 2.000000 1.000000 0.100000
25% 5.100000 2.800000 1.600000 0.300000
50% 5.800000 3.000000 4.350000 1.300000
75% 6.400000 3.300000 5.100000 1.800000
max 7.900000 4.400000 6.900000 2.500000
# Summary by group
DF.groupby('Species').size()Species
setosa 50
versicolor 50
virginica 50
dtype: int64
30.4 Train-Test Split 
# Train-Test split
train, test = train_test_split(DF, test_size = 0.3, stratify = DF[['Species']], random_state = 1234)
print('Full Data: ', DF.shape)Full Data: (150, 5)print('Train: ', train.shape)Train: (105, 5)print('Test: ', test.shape)Test: (45, 5)
30.5 Cross-validation 
We can employ a procedure called cross-validation (CV) using the training data rather than defining a train and validation split. This provides a more effective way to determine the generalisation performance when comparing models or parameters. The following implementation is depicted here:
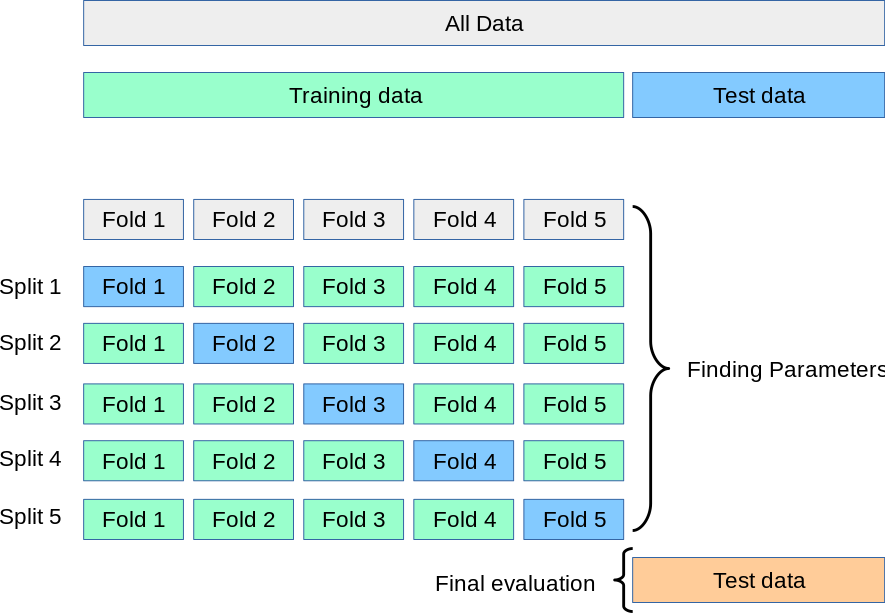
Five-fold cross-validation strategy
Image credit: scikit-learn
Import the ShuffleSplit function which allows you to define the number of folds, and the proportion of data for testing, in this case 30%.
from sklearn.model_selection import ShuffleSplit
cv = ShuffleSplit(n_splits=5, test_size=0.3, random_state=0)Once you have defined the splits, you can pass this to the cross_val_score which is described in section on fitting the classification model with cross-validation.
30.6 Exploratory Data Analysis 
Note: The code demonstrates only a selective exploratory data analysis; it is not comprehensive.
# Scatter plot matrix
sns.pairplot(train, hue = 'Species', palette = 'colorblind')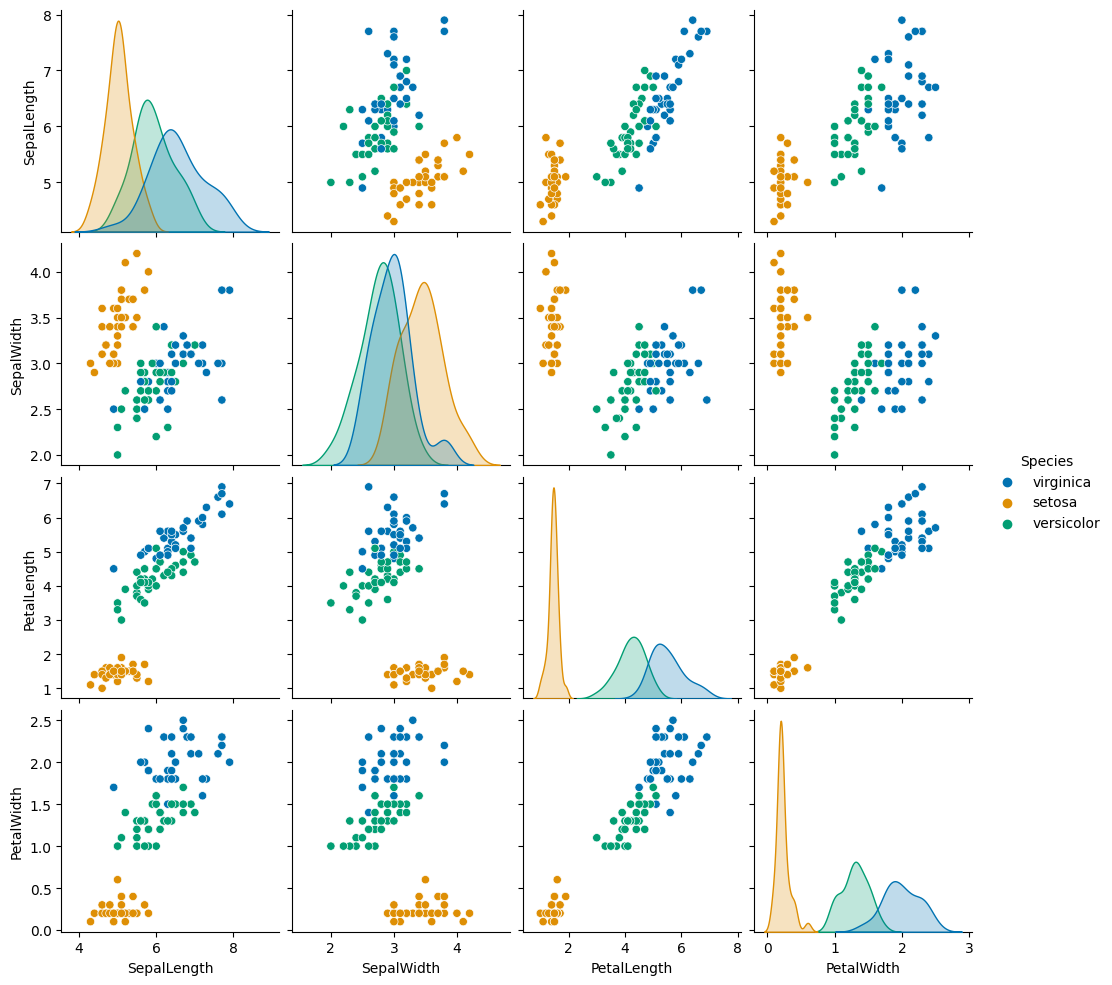
# Correlation plot
plt.clf()
corrmat = train.corr()
sns.heatmap(corrmat, annot = True, square = True);
plt.show()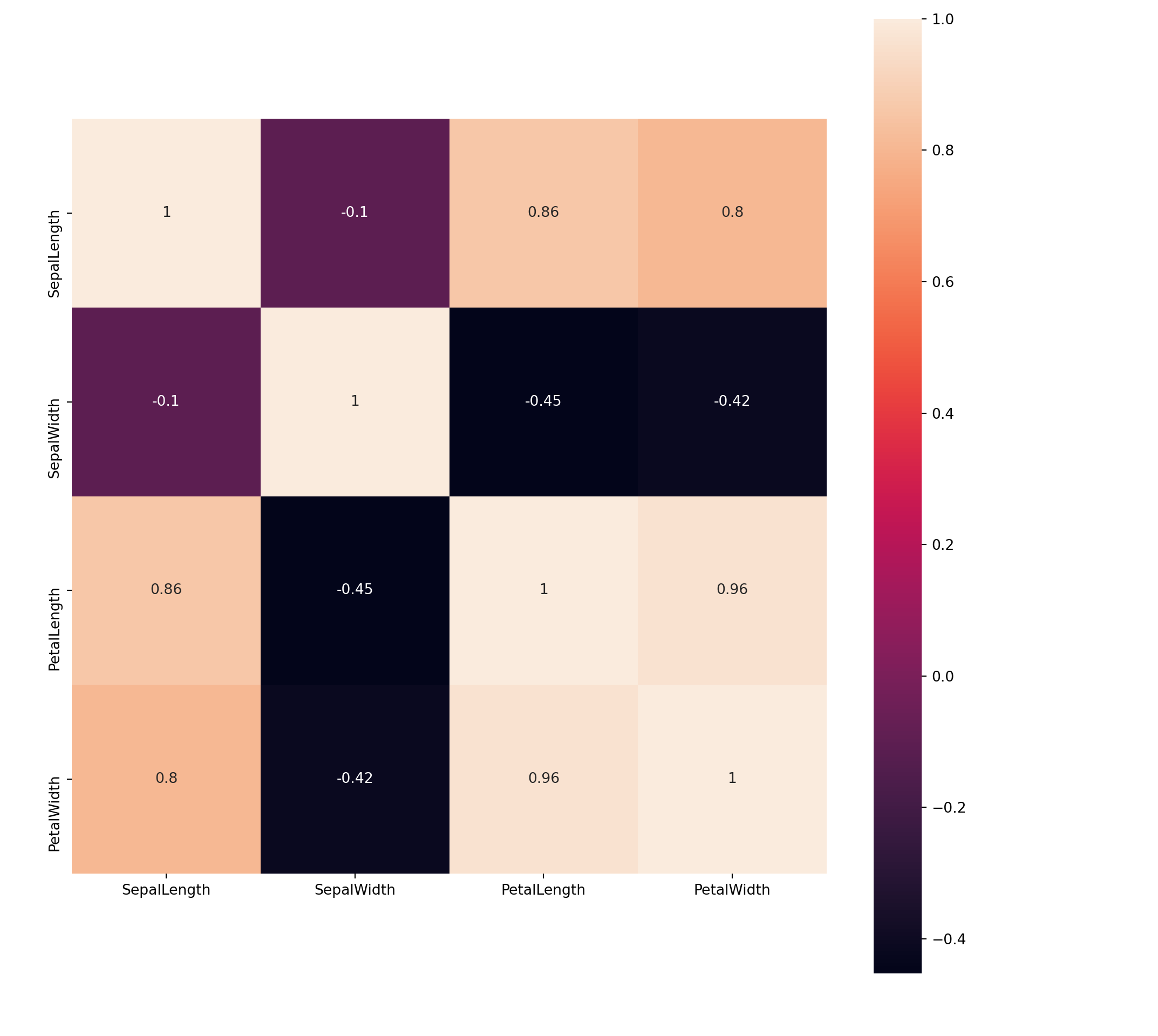
30.6.1 Build and fit the classification model 
Here, we build a Decision Tree classifier
X_train = train.drop(['Species'], axis = 1)
y_train = train[['Species']]
X_test = test.drop(['Species'], axis = 1)
y_test = test[['Species']]
# feature names
fn = X_train.columns
# class names
cn = ['setosa', 'versicolor', 'virginica']
clf_DT = DecisionTreeClassifier(max_depth = 3, random_state = 123)
clf_DT.fit(X_train, y_train)DecisionTreeClassifier(max_depth=3, random_state=123)Note: Python allows all assignments in a single line
X_train, y_train, X_test, y_test = train.drop(['Species'], axis = 1), train[['Species']], test.drop(['Species'], axis = 1), test[['Species']]
30.6.2 Test the model on Test data 
Note
test_pred provides the class names
test_prob provides the probability for each class names
test_pred = clf_DT.predict(X_test)
test_prob = clf_DT.predict_proba(X_test)
print('Test Prediction: ', test_pred.shape)Test Prediction: (45,)
30.6.3 Model Performance Metrics 
print('Model Accuracy: ', f'{metrics.accuracy_score(test_pred, y_test): 0.4f}')Model Accuracy: 0.9556print("Precision, Recall, Confusion matrix, in training\n")
# Precision Recall scoresPrecision, Recall, Confusion matrix, in trainingprint(metrics.classification_report(y_test, test_pred, digits = 3))
# Confusion matrix precision recall f1-score support
setosa 1.000 1.000 1.000 15
versicolor 0.933 0.933 0.933 15
virginica 0.933 0.933 0.933 15
accuracy 0.956 45
macro avg 0.956 0.956 0.956 45
weighted avg 0.956 0.956 0.956 45print(metrics.confusion_matrix(y_test, test_pred))[[15 0 0]
[ 0 14 1]
[ 0 1 14]]
plt.clf()
CM = metrics.plot_confusion_matrix(clf_DT, X_test, y_test, normalize = None)
CM.ax_.set_title('Decision Tree Confusion matrix, without normalization');
# plt.show()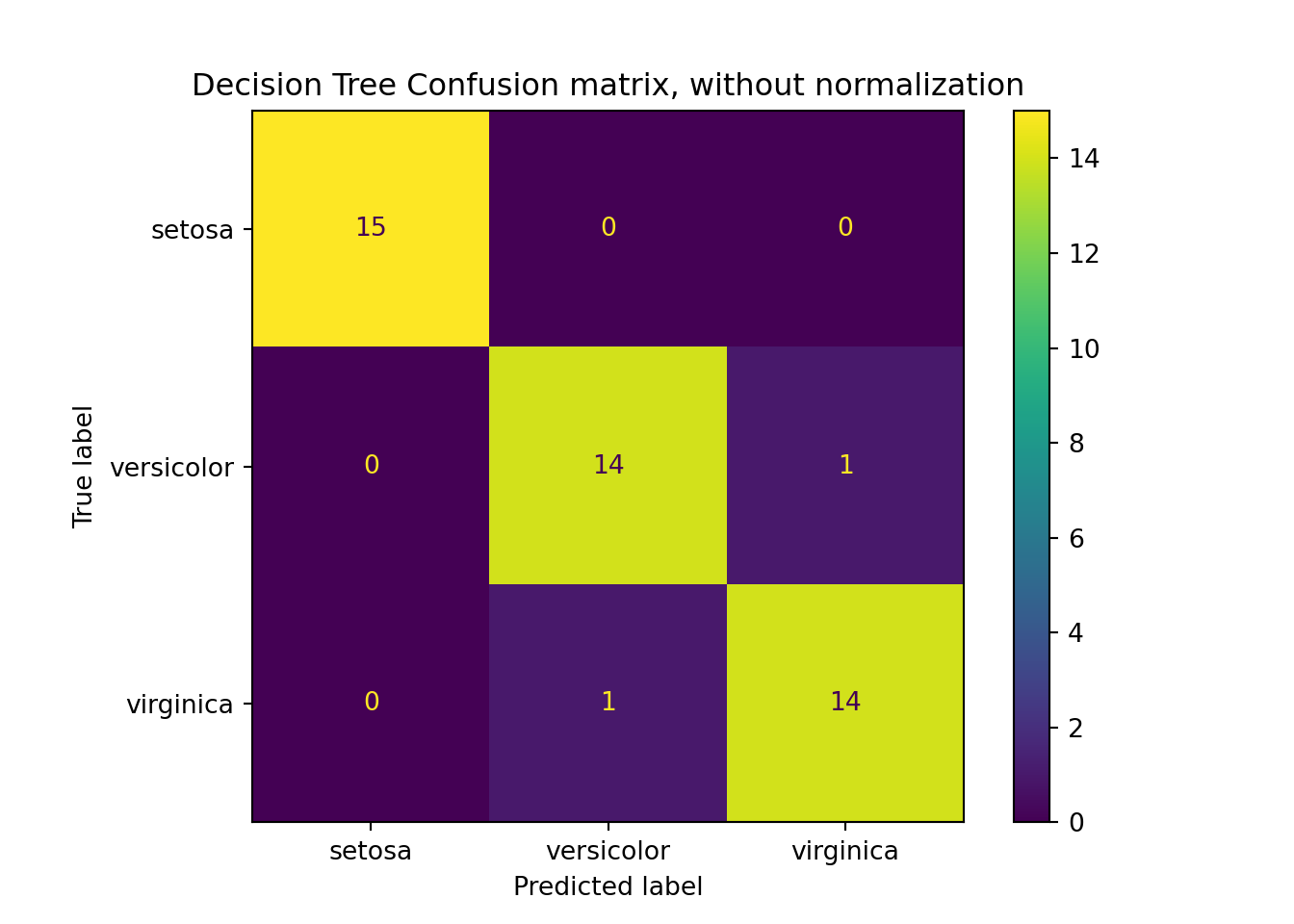
Fit the classification model with cross validation
To employ cross validation on our previously defined folds, simply import the cross_val_score function and append the following lines after you have fit the model.
from sklearn.model_selection import cross_val_score
scores = cross_val_score(clf_DT, x_train, y_train, cv=cv, scoring='accuracy')This will produce 5 accuracy performance metrics per fold, which we configured to be 5.