Section 42 Post hoc analysis: R implementation
42.1 Familywise Error Rate
Analysis of Variance Table
Response: SBP
Df Sum Sq Mean Sq F value Pr(>F)
Group 3 1521.6 507.21 7.4983 0.0005059 ***
Residuals 36 2435.2 67.64
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Pairwise comparisons using t tests with pooled SD
data: BP$SBP and BP$Group
A B C
B 0.0292 - -
C 0.2067 0.3313 -
D 5.3e-05 0.0266 0.0022
P value adjustment method: none # Multiple comparison: Bonferroni correction
pairwise.t.test(x = BP$SBP, g = BP$Group, p.adjust.method = "bonferroni")
Pairwise comparisons using t tests with pooled SD
data: BP$SBP and BP$Group
A B C
B 0.17548 - -
C 1.00000 1.00000 -
D 0.00032 0.15975 0.01325
P value adjustment method: bonferroni
Pairwise comparisons using t tests with pooled SD
data: BP$SBP and BP$Group
A B C
B 0.10650 - -
C 0.41333 0.41333 -
D 0.00032 0.10650 0.01104
P value adjustment method: holm Tukey multiple comparisons of means
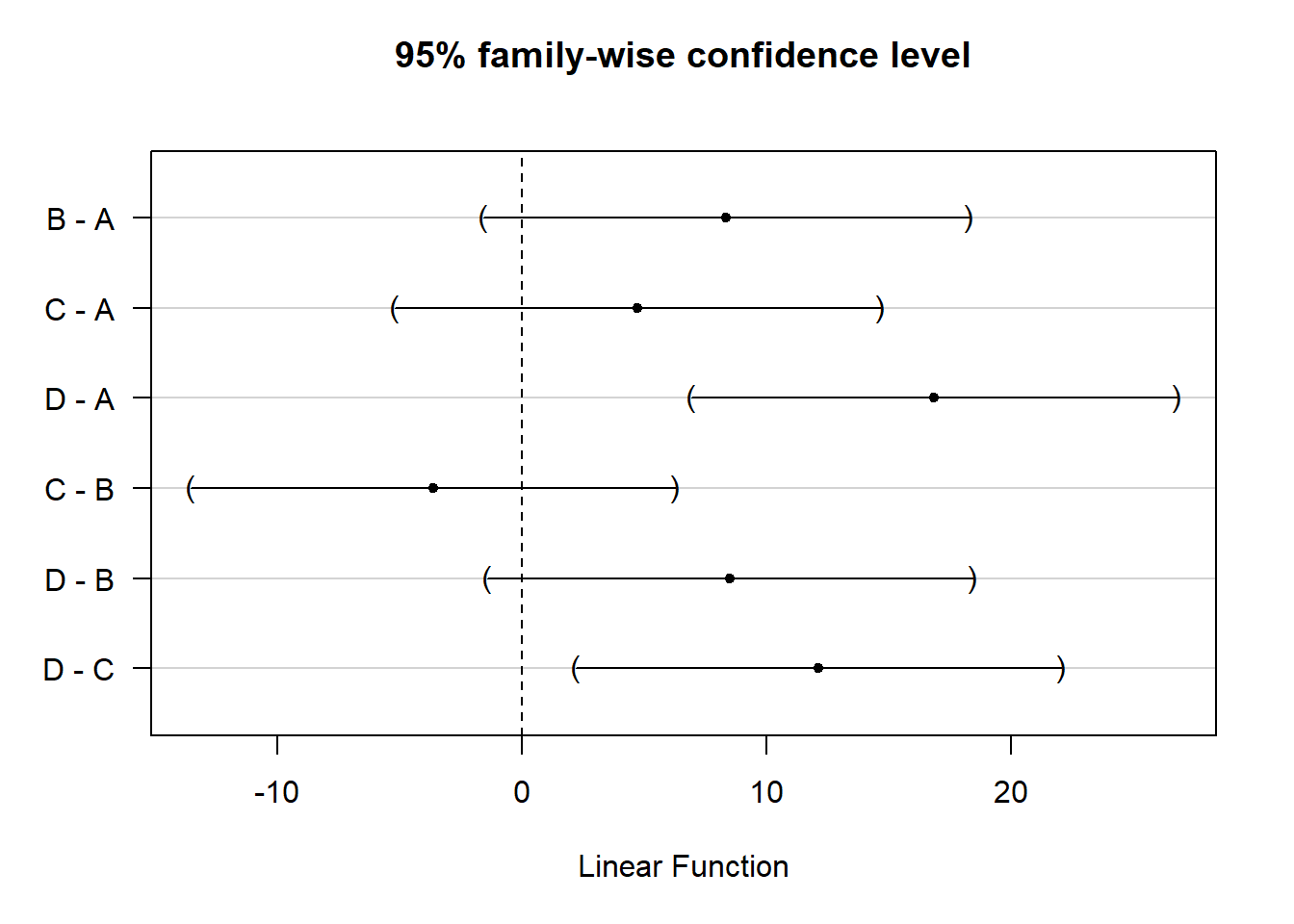
95% family-wise confidence level
Fit: aov(formula = SBP ~ Group, data = BP)
$Group
diff lwr upr p adj
B-A 8.352 -1.554098 18.258098 0.1240483
C-A 4.730 -5.176098 14.636098 0.5775956
D-A 16.855 6.948902 26.761098 0.0003004
C-B -3.622 -13.528098 6.284098 0.7589637
D-B 8.503 -1.403098 18.409098 0.1142036
D-C 12.125 2.218902 22.031098 0.0113324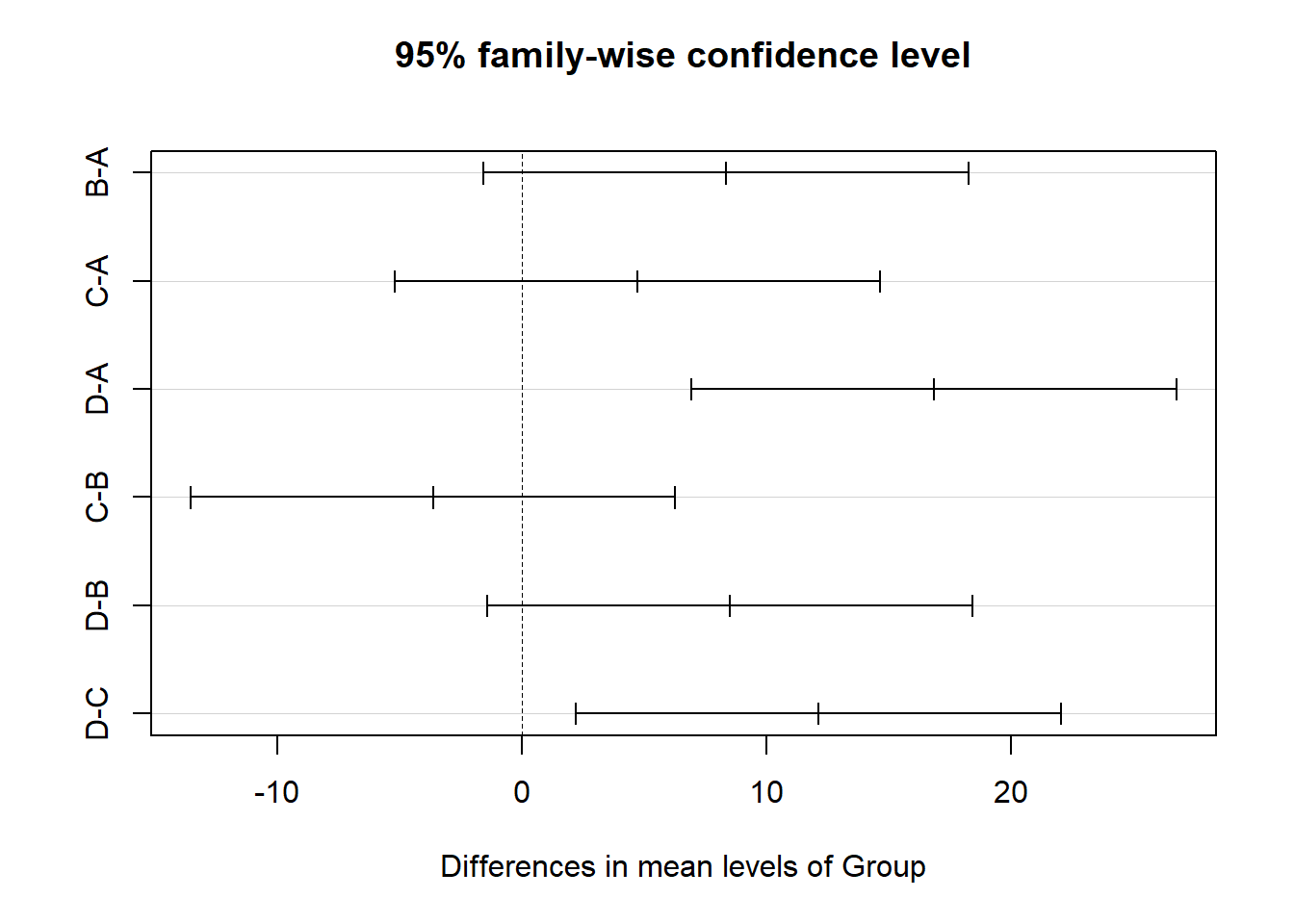
42.2 False Discovery Rate
# Benjamini & Hochberg (1995) BH or fdr
pairwise.t.test(x = BP$SBP, g = BP$Group, p.adjust.method = "BH")
Pairwise comparisons using t tests with pooled SD
data: BP$SBP and BP$Group
A B C
B 0.04387 - -
C 0.24800 0.33133 -
D 0.00032 0.04387 0.00662
P value adjustment method: BH
Pairwise comparisons using t tests with pooled SD
data: BP$SBP and BP$Group
A B C
B 0.10748 - -
C 0.60759 0.81175 -
D 0.00079 0.10748 0.01623
P value adjustment method: BY 42.3 Using library(lsmeans)
require(lsmeans)
fm <- lm(SBP ~ Group, data = BP)
gr.means <- lsmeans(fm, specs = ~Group)
# Esimated means
gr.means Group lsmean SE df lower.CL upper.CL
A 98.9 2.6 36 93.7 104
B 107.3 2.6 36 102.0 113
C 103.7 2.6 36 98.4 109
D 115.8 2.6 36 110.5 121
Confidence level used: 0.95 contrast estimate SE df t.ratio p.value
A - B -8.35 3.68 36 -2.271 0.0292
A - C -4.73 3.68 36 -1.286 0.2067
A - D -16.86 3.68 36 -4.582 0.0001
B - C 3.62 3.68 36 0.985 0.3313
B - D -8.50 3.68 36 -2.312 0.0266
C - D -12.12 3.68 36 -3.296 0.0022# Comparison of means: FWER (bonferroni)
contrast(gr.means, method = "pairwise", adjust = "bonferroni") contrast estimate SE df t.ratio p.value
A - B -8.35 3.68 36 -2.271 0.1755
A - C -4.73 3.68 36 -1.286 1.0000
A - D -16.86 3.68 36 -4.582 0.0003
B - C 3.62 3.68 36 0.985 1.0000
B - D -8.50 3.68 36 -2.312 0.1597
C - D -12.12 3.68 36 -3.296 0.0132
P value adjustment: bonferroni method for 6 tests contrast estimate SE df t.ratio p.value
A - B -8.35 3.68 36 -2.271 0.1240
A - C -4.73 3.68 36 -1.286 0.5776
A - D -16.86 3.68 36 -4.582 0.0003
B - C 3.62 3.68 36 0.985 0.7590
B - D -8.50 3.68 36 -2.312 0.1142
C - D -12.12 3.68 36 -3.296 0.0113
P value adjustment: tukey method for comparing a family of 4 estimates contrast estimate SE df t.ratio p.value
A - B -8.35 3.68 36 -2.271 0.0439
A - C -4.73 3.68 36 -1.286 0.2480
A - D -16.86 3.68 36 -4.582 0.0003
B - C 3.62 3.68 36 0.985 0.3313
B - D -8.50 3.68 36 -2.312 0.0439
C - D -12.12 3.68 36 -3.296 0.0066
P value adjustment: fdr method for 6 tests 42.4 Using library(multcomp)
The package multcomp considers a wide range of methods as well as customised contrasts to conduct multiple testing of hypotheses.
require(multcomp)
# Prepare the contrast using Group='Tukey'
mult.test <- glht(fm, linfct = mcp(Group = "Tukey"))
# FWER: tukey
summary(mult.test)
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: Tukey Contrasts
Fit: lm(formula = SBP ~ Group, data = BP)
Linear Hypotheses:
Estimate Std. Error t value Pr(>|t|)
B - A == 0 8.352 3.678 2.271 0.1239
C - A == 0 4.730 3.678 1.286 0.5776
D - A == 0 16.855 3.678 4.582 <0.001 ***
C - B == 0 -3.622 3.678 -0.985 0.7590
D - B == 0 8.503 3.678 2.312 0.1142
D - C == 0 12.125 3.678 3.296 0.0112 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Adjusted p values reported -- single-step method)
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: Tukey Contrasts
Fit: lm(formula = SBP ~ Group, data = BP)
Linear Hypotheses:
Estimate Std. Error t value Pr(>|t|)
B - A == 0 8.352 3.678 2.271 0.02925 *
C - A == 0 4.730 3.678 1.286 0.20666
D - A == 0 16.855 3.678 4.582 5.34e-05 ***
C - B == 0 -3.622 3.678 -0.985 0.33133
D - B == 0 8.503 3.678 2.312 0.02662 *
D - C == 0 12.125 3.678 3.296 0.00221 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Adjusted p values reported -- none method)
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: Tukey Contrasts
Fit: lm(formula = SBP ~ Group, data = BP)
Linear Hypotheses:
Estimate Std. Error t value Pr(>|t|)
B - A == 0 8.352 3.678 2.271 0.17548
C - A == 0 4.730 3.678 1.286 1.00000
D - A == 0 16.855 3.678 4.582 0.00032 ***
C - B == 0 -3.622 3.678 -0.985 1.00000
D - B == 0 8.503 3.678 2.312 0.15975
D - C == 0 12.125 3.678 3.296 0.01325 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Adjusted p values reported -- bonferroni method)
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: Tukey Contrasts
Fit: lm(formula = SBP ~ Group, data = BP)
Linear Hypotheses:
Estimate Std. Error t value Pr(>|t|)
B - A == 0 8.352 3.678 2.271 0.04387 *
C - A == 0 4.730 3.678 1.286 0.24800
D - A == 0 16.855 3.678 4.582 0.00032 ***
C - B == 0 -3.622 3.678 -0.985 0.33133
D - B == 0 8.503 3.678 2.312 0.04387 *
D - C == 0 12.125 3.678 3.296 0.00662 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Adjusted p values reported -- fdr method)
Simultaneous Confidence Intervals
Multiple Comparisons of Means: Tukey Contrasts
Fit: lm(formula = SBP ~ Group, data = BP)
Quantile = 2.6954
95% family-wise confidence level
Linear Hypotheses:
Estimate lwr upr
B - A == 0 8.3520 -1.5619 18.2659
C - A == 0 4.7300 -5.1839 14.6439
D - A == 0 16.8550 6.9411 26.7689
C - B == 0 -3.6220 -13.5359 6.2919
D - B == 0 8.5030 -1.4109 18.4169
D - C == 0 12.1250 2.2111 22.0389