Section 34 ggplot2 scale
34.1 scale
All functions with prefix scale_ control how a plot maps data values to the visual values of an aesthetic. To change the ggplot2 default mapping, you need to add a custom scale.
Override the default scales to tweak details like the axis labels or legend keys, or to use a completely different translation from data to aesthetic. labs() and lims() are convenient helpers for the most common adjustments to the labels and limits.
Find more details here: http://ggplot2.tidyverse.org/reference/

34.2 aesthetics
| aesthetics | Description |
|---|---|
colour |
Color of lines and points |
fill |
Color of area fills (e.g. bar graph) |
linetype |
Solid/dashed/dotted lines |
shape |
Shape of points |
size |
Size of points |
alpha |
Opacity/transparency |
x |
X-axis |
y |
Y-axis |
34.3 General purpose scale
| type | Description |
|---|---|
manual |
Map discrete values to manually-specified values (e.g., colors, point shapes, line types) |
discrete |
Map discrete values to visual values (e.g., colors, point shapes, line types, point sizes) |
continuous |
Map continuous values to visual values (e.g., alpha, colors, point sizes) |
identity |
Use data values as visual values |
There are other scale types that control date, time, colour, scale of axes, shape and area. Check the ggplot2 website for further details.
34.4 scale-specific arguments
Arguments corresponding to a type of scale
Common arguments are: values, limits, name, labels, palette
34.5 Default boxplot
g <- ggplot(data=iris,
mapping=aes(x=Species, y=Sepal.Length, fill=Species))
g <- g + geom_boxplot()
g <- g + labs(title='Boxplot of iris data',
subtitle = 'Based on iris data',
x='Species',
y='Sepal Length (cm)')
g <- g + theme_bw()
g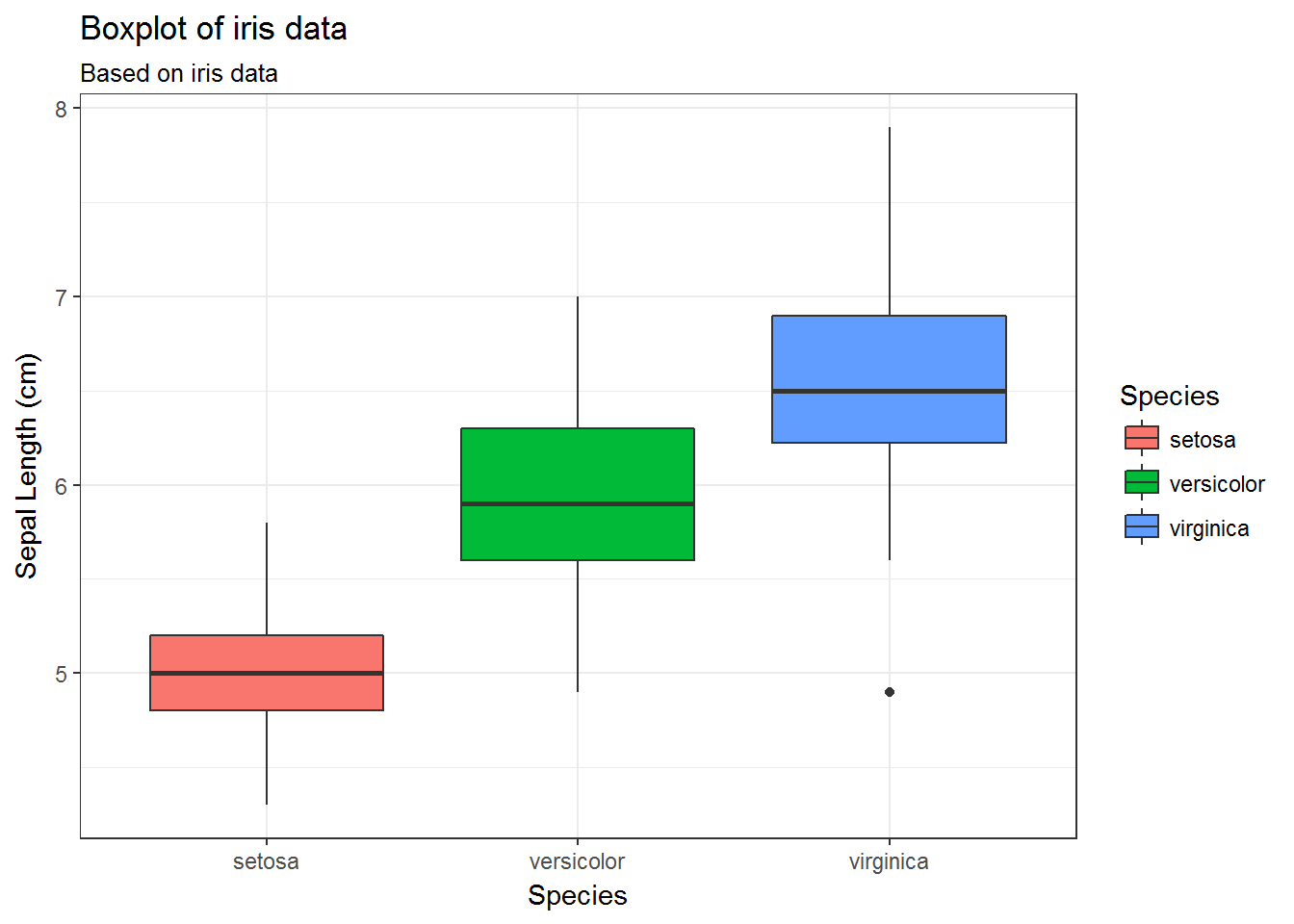
34.6 Remove legends using scale_
# Option 3: Remove legend specifying the scale
g + scale_fill_discrete(guide=FALSE)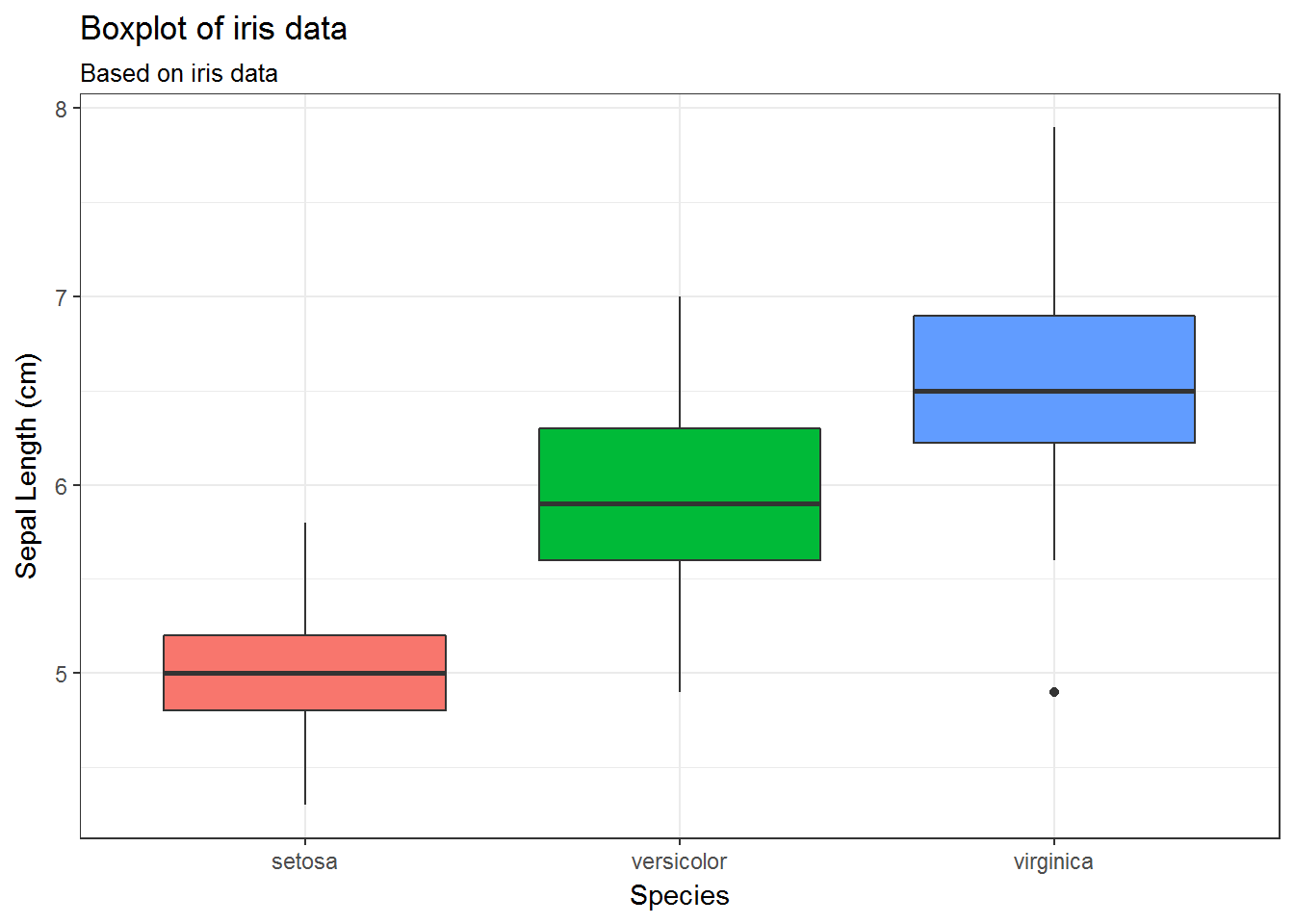
34.7 Modify legend titles and labels
g + scale_fill_discrete(name='Iris Species')
g + scale_fill_discrete(name='Iris Species',
breaks=c('setosa', 'versicolor', 'virginica'),
labels=c('Setosa', 'Versicolor', 'Virginica'))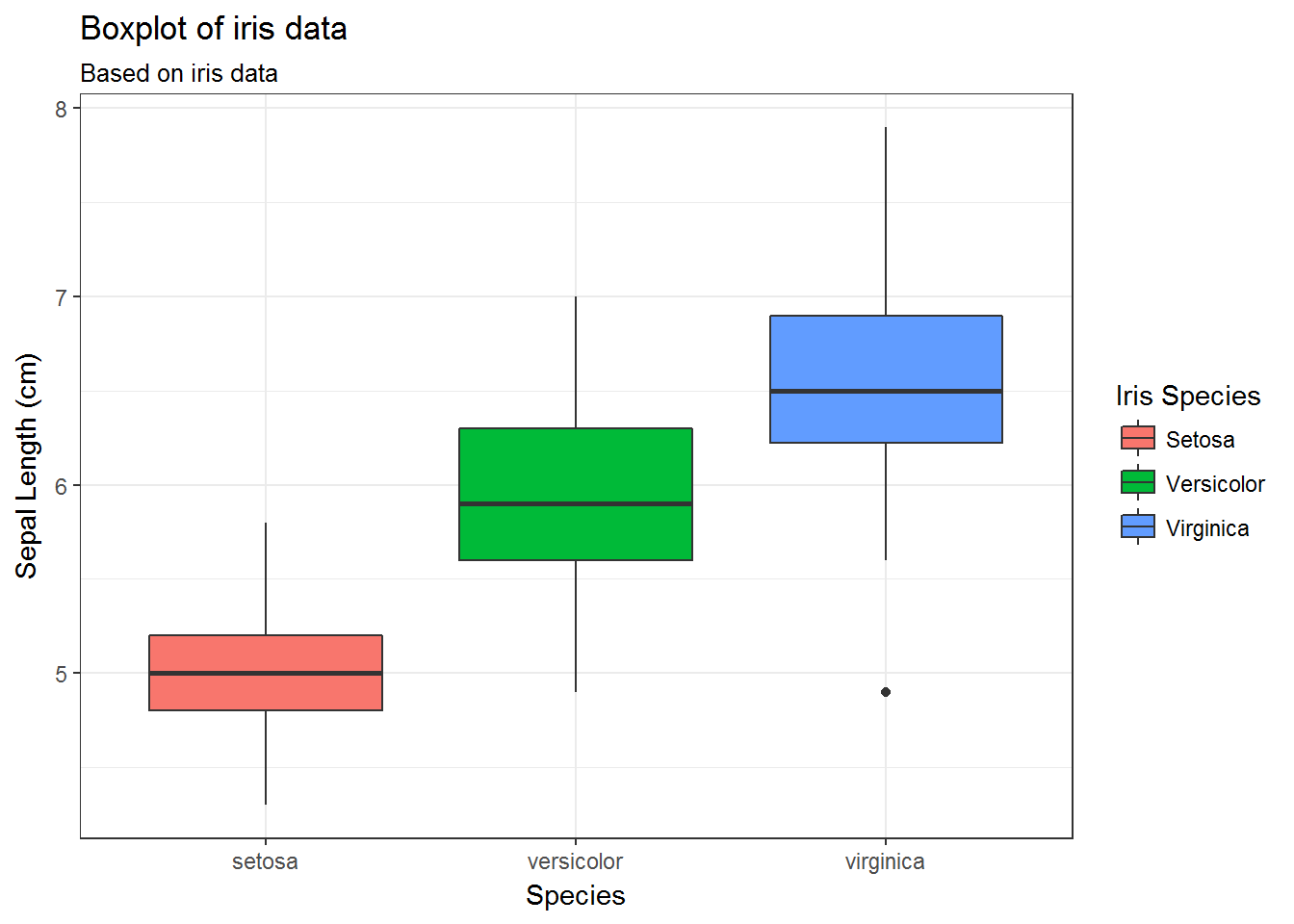
34.8 scale_fill_manual
g <- ggplot(data=iris,
mapping=aes(x=Species, y=Sepal.Length, fill=Species))
g <- g + geom_boxplot()
g <- g + stat_summary(fun.y=mean, geom='point', shape=16, size=4, colour='yellow')
g + scale_fill_manual(values=c('blue', 'pink', 'green'),
name='Iris Species',
breaks=c('setosa', 'versicolor', 'virginica'),
labels=c('Setosa', 'Versicolor', 'Virginica'))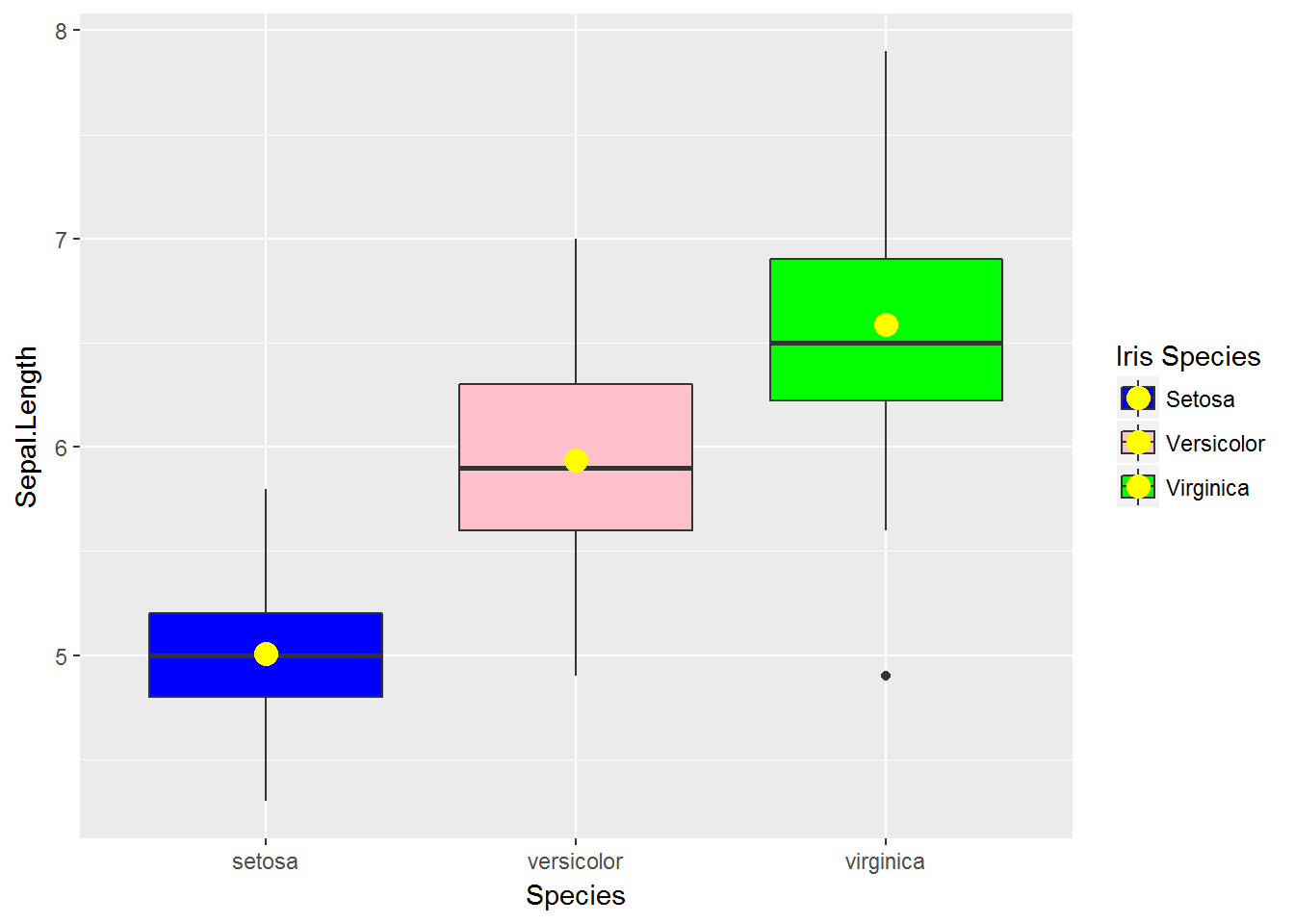
34.9 Change both x-axis and legend labels
g + scale_fill_manual(values=c('blue', 'pink', 'green'),
name='Iris Species',
breaks=c('setosa', 'versicolor', 'virginica'),
labels=c('Setosa', 'Versicolor', 'Virginica')) +
scale_x_discrete(breaks=c('setosa', 'versicolor', 'virginica'),
labels=c(c('Setosa', 'Versicolor', 'Virginica')))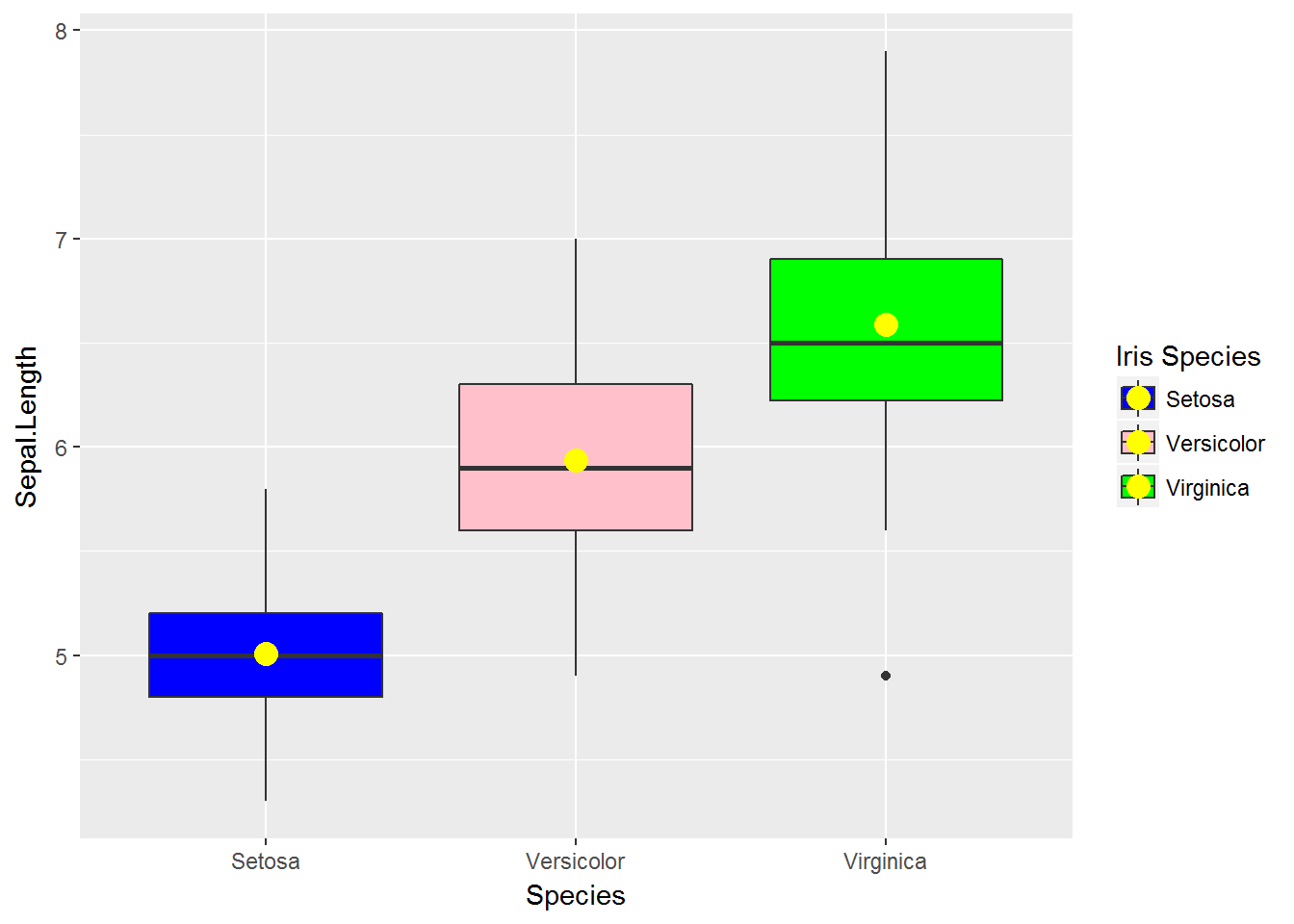
34.10 Change the data.frame
An alternative (and easier) way to show the appropriate labels of a factor in the ggplot2 outputs, first change the labels of the factor in the data.frame, and then call ggplot2 functions.
mod.iris <- iris
mod.iris$Species <- factor(mod.iris$Species,
levels = c('setosa', 'versicolor', 'virginica'),
labels = c('Setosa', 'Versicolor', 'Virginica'))
g <- ggplot(data=mod.iris,
mapping=aes(x=Species, y=Sepal.Length, fill=Species))
g <- g + geom_boxplot()
g <- g + labs(title='Boxplot of iris data',
subtitle = 'Based on iris data',
x='Species',
y='Sepal Length (cm)')
g <- g + theme_bw()
g
34.11 Exercise
- Generate the following data.
set.seed(12345)
x <- rnorm(n = 10, mean=10, sd=2)
y1 <- 2*x + rnorm(n = 10, mean=0, sd=1)
y2 <- 4 + 2*x + rnorm(n = 10, mean=0, sd=1)
Gr <- rep(c('G1','G2'), each = 10)
DF <- data.frame(X=x, Y=c(y1,y2), ID=LETTERS[1:20], Gr=Gr)- Create the following scatter plot along with the fitted line. Change the colour of points as red and blue for Gr1 and Gr2, respectively.
